Karnataka 2nd PUC Biology Important Questions Chapter 6 Molecular Basis of Inheritance
Question 1.
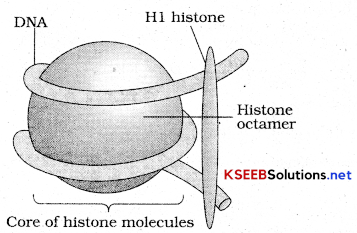
Explain the structure of Nucleosome
Answer:
In prokaryotes, such as E coli through they do not have a defined nucleus the DNA is not scattered throughout the cell. DNA being negatively charged is held with some proteins that have positive charges in a region termed as nucleoid DNA in nucleoid is organized in large loops held by proteins.
In eukaryotes this organization is more complex. There is a set of positively changed basic proteins termed as histones. Histones are rich in amino acids arginine and lysine, which carry the charges in their side chains. There is an octamer of histone mojcehle and negatively charged DNA is wrapped and histone octamer is formed.
Nucleosome: A typical nucleosome contains 200bp of DNA helix. The nucleosomes in chromatin are seen as beads on string when seen in electron microscope. In a typical nucleus some regions of chromatin are loosely packed and light and are termed as euchromatin. Densely packed region of chromatin are heterochromatin which are dark in color. Euchromatin is active and heterochromatin is inactive, transcriptionally.
Question 2.
Name a nitrogenous base found only in DNA but absent in RNA?
Answer:
Thymine.
Question 3.
Name the nitrogenous base present in RNA but absent in DNA?
Answer:
Uracil.
![]()
Question 4.
Define nucleoside?
Answer:
A nucleoside is a combination of base and sugar.
Question 5.
Define nucleotide?
Answer:
It is a combination of Base, Sugar and phosphate.
Question 6.
What is Chargaff’s base equivalent rule?
Answer:
Total number of purines is equal to the total number of pyrimidine’s and their ratio is constant.
Question 7.
What do you mean by euchromatin?
Answer:
Lightly stained chromatin is euchromatin.
Question 8.
What do you mean by heterochromatin?
Answer:
Darkly stained chromatin is heterochromatin.
Question 9.
List the purines and pyrimidines in DNA?
Answer:
Purines Pyrimidines
Adenine and Guanine Cytosine Thymine.
Question 10.
List the nucleosides in DNA?
Answer:
- Deoxyadenosine
- Deoxyguanosine
![]()
Question 11.
List the nucleotides in DNA?
Answer:
- Deoxyadenylic acid
- Deoxyguanylic acid
- Deoxythymidylic acid
- Deoxycytidylic acid.
Question 12.
List the nucleosides of RIN A?
Answer:
- Adenosine,
- Guanosine
- Uridine
- Cytidine,
Question 13.
List the nucleotides of RNA?
Answer:
- Adenylic acid
- Guanylic acid.
- Uridylic acid
- Cytidylic acid.
Question 14.
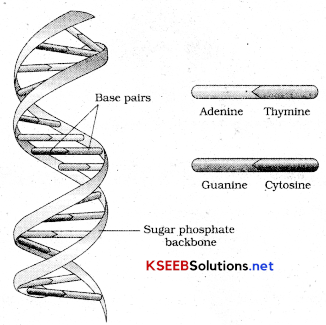
Describe the double-helical structure of DNA with a labeled diagram?
Answer:
Question 15.
What is a nucleosome? Explain the structure of nucleosome with a neat labeled diagram?
Answer:
In prokaryotes, such as E coli through they do not have a defined nucleus the DNA is not scattered throughout the cell. DNA being negatively charged is held with some proteins that have positive charges in a region termed as nucleoid DNA in nucleoid is organized in large loops held by proteins.
In eukaryotes this organization is more complex. There is a set of positively changed basic proteins termed as histones. Histones are rich in amino acids arginine and lysine, which carry the charges in their side chains. There is an octamer of histone mojcehle and negatively charged DNA is wrapped and histone octamer is formed.
Nucleosome: A typical nucleosome contains 200bp of DNA helix. The nucleosomes in chromatin are seen as beads on string when seen in electron microscope. In a typical nucleus some regions of chromatin are loosely packed and light and are termed as euchromatin. Densely packed region of chromatin are heterochromatin which are dark in color. Euchromatin is active and heterochromatin is inactive, transcriptionally.

Question 16.
How did Avery, Macleod and Mccarty proved biochemical nature of transforming principle?
Answer:
Avery, Maccleod and Mccavty’s work stated the genetic material was thought to be a protein. They purified biochemical (Proteins, DNA, RNA etc) from the heat killed S cells to see which ones could transform R cells into S cells. They discovered that DNA from S bacteria caused R bacteria to become transformed.
They also discovered that protein digesting enzymes (proteases) and RNA digesting enzymes (RNases) did not affect transformation so the transforming substance was not protein or RNA. Digestion with DNase inhibited transformation suggesting that DNA caused the transformation.
Thus DNA is the hereditary material in all organisms.
![]()
Question 17.
Differentiate eukaryotic genes from prokaryotic genes?
Answer:
| 1. Eukaryotic gene are splits genes | 1. Prokaryotic genes contain only exons contain both introns and exons |
| 2. RNA Polymerase is of 3 types in Eukaryotic | 2. It is of type only in prokaryotes Gene |
| 3. It produces monocistronic m RNA | 3. It produces polycistronic m RNA |
Question 18.
Describe Griffith’s experiment on transformation to prone DNA is the genetic material?
Answer:
Question 19.
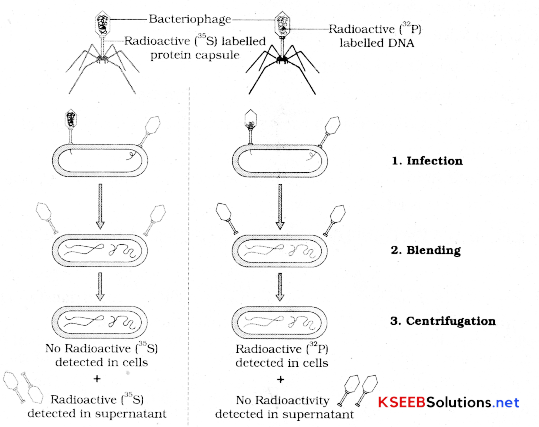
Describe Hershey and Chase’s experiment to prove DNA is the genetic material?
Answer:
Alfred Hershey and Martha Chase also gave experiments to prove DNA is the genetic material. They worked on viruses infecting bacteria called bacteriophages.
Bacteriophage attaches to bacteria and its genetic material enters bacterial cell. The bacterial cell treats the viral genetic material as if it was its own and manufactures more virus particles.
Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria.
They grow some viruses on medium that contained radioactive phosphorus and some others on medium that contained radioactive sulfur.
Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not.
Similarly viruses growing on radioactive sulfur contained radioactive protein but not radioactive DNA because DNA does not contain sulfur.
Radioactive phage were allowed to attach to E. coli bacteria. As the infection proceeded viral coats were removed from the bacteria agitating them in ablender. Virus particles were separated from bacteria by spinning them in a centrifuge.
Bacteria infected with viruses that had radioactive DNA were radioactive indicating that DNA was the material that passed from virus to bacteria.
Bacteria that were infected with viruses that had radioactive proteins were not radioactive. This indicates that proteins did not enter the bacteria from the viruses DNA is the enetic material that is. passed from virus to bacteria.
Question 20.
What are split genes?
Answer:
Eukaryotic genes are termed as split genes as they contain both introns and exons.
![]()
Question 21.
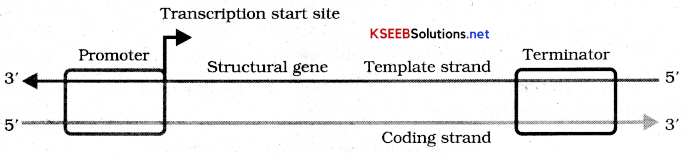
Define transcription. Describe the process of transcription in prokaryotes?
Answer:
Transcription Unit: A transcription unit in DNA is defined primarily by three regions in DNA.
- A promoter
- Structural gene
- A terminator
Since the two strands have opposite polarity and the DNA dependent RNA polymerase also called as polymerization in only one direction that is 5’ – 3’ the strand that has polarity 3’ – 5’ – acts a template and is referred to as template strand. The strand which does not code for anything is coding strand.
The promoter and terminator flank the structural gene in transcription unit. The promoter is said to be located towards 5’ end of the structural gene. It is a DNA sequence that provides binding site for RNA polymerase. Terminator is located towards 3 ’ end of the coating strand and it defines the end of the process of transcription.
Transcription Unit and Gene – Cistron is a segment of DNA Coding for a Polypeptide. Structural gene in a transcription unit could be monocistronic (mostly in eukaryotes) or polycistronic(bacteria or prokaryotes)
In Eukaryotes monocistronic’ structural genes are split. Coding sequences are defined as exons. Exons are interrupted by introns. Introns do not appear in mature or processed RNA.
Question 22.
Describe mechanism of semiconservative DNA replication?
Answer:
DNA replication: The parental DNA molecule produces two identical copies of daughter DNA molecules and it is called DNA replication.
- The process of replication begins at a specific point on DNA strand called initiation site or site of origin (ORI).
- The hydrogen bonds present between two strands are dissolved by DNA polymerase I.
- The double stranded DNA is now getting separated to form single strands.
This mechanism is called unzipping of DNA. - As the double helix is separated it gets into Y shaped appearance called replication fork. Consequently on one strand (the template with polarity (3’ – 5’) the replication is continuous while on the other (polarity 5’ – 3 ’) it is discontinuous. Discontinuously synthesized fragments are joined together by DNA ligase.
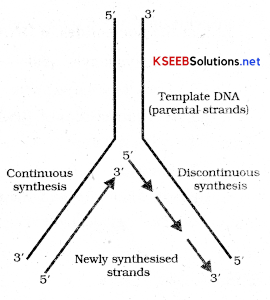
With reference to transcription define:
(a) Splicing – In enparyotes pritranscripts contain both exons and introns. Introns is non functional and it is subjected to splicing.
(b) Capping – Introns are removed and oxensone joined in a definite order in part.
Question 23.
Expand VNTR?
Answer:
Variable Number of Tandem Repeats.
Question 24.
Which is the largest human gene?
Answer:
Dystrophin.
Question 25.
Why t RNA is termed as adaptor molecule?
Answer:
It helps in transfer of amino acids to the site of protein synthesis hence it is termed as an adaptor molecule.
Question 26.
Name the initiator codon and terminator codons?
Answer:
AUG is termed as initiator codon and terminator codons are UAA, UAG, UGA.
![]()
Question 27.
Explain any two properties of genetic code
Answer:
- Genetic code is universal: It is common and found in all living organisms.
- Triplet codon: Genetic code is a triplet code and is represented by three-nucleotide sequences present on m RNA strand.
- Initiator codon is AUG which codes for methionine. In case AUG is absent GUG acts as an initiator Codon which codes for amino acid valine.
Question 28.
Give any four important characteristic features of genetic code?
Answer:
Features:
- Genetic code is universal: It is common and found in all living organisms.
- Triplet codon: Genetic code is a triplet code and is represented by three nucleotide sequences present on m RNA strand.
- Initiator codon is AUG which codes for methionine. In case AUG is absent GUG acts as an initiator Co don which codes for amino acid valine.
- The process of translation is stopped by terminator Codons UAA, UAG, UGA which do not code for any amino acid.
- Draw a neat labeled diagram of t RNA molecule.
Question 29.
Explain process of translation in detail.
Answer:
Translation: The process of polymerization of amino acids to form a polypeptide is termed as translation. The process of translation takes place in the cytoplasm of the cell where ribosome is located and it takes place in 4 levels
1. Activation or charging of amino acid: Amino acids before they attach to t RNA should be activated by using ATP molecules.
Amino acid + ATP- Activated amino acid.
Activated amino acid will be picked up by at RNA molecule and form amino a CY1 tRNA.
This process is called charging of amino acid.
2. Chain initiation:
- The formation of polypeptide chain is initiated by the initiator codon AUG.
- First mRNA binds to smaller subunit of ribosome ie (3os) and later on larger submit joins.
- Larger sub unit (5os) has a site (Aminoacyl site), P site peptidyl site (enzyme peptidyl transfer are and an E site or exit site.
Image
3. Chain elongation: Ribosomal subunit shifts from one codon to another codon on the m RNA strand. T RNA carrying amino acid shifts from A site to P site. At P site of 5os subunit peptide bond formation takes place between 2 amino acids and it is catalyzed by peptidyl transfer enzyme and elongation factos EF , EF„ EF,. chain keeps elongating.
4. Chain termination: Codons present on m RNA strand are translated by ribosomes continuously to form a long polypeptide chain until the ribosomes reach the terminator codons (UAA, UAG, UGA).
Question 30.
With a neat labeled diagram explain Lac operon concept.
Answer:
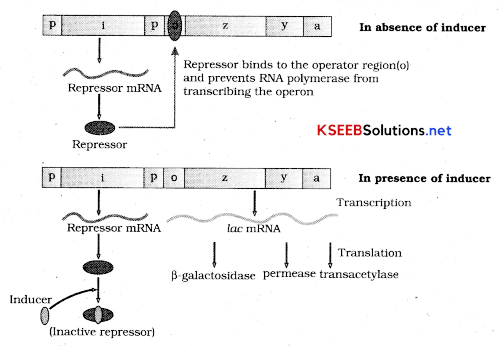
Lac operon concept: The lac operon concept was given by Jacob and Monod. This concept was found in bacteria.
Lac operon consists of one regulatory’ gene (I gene) and three structural gene (Z, y; a) promoter gene gene Z codes for enzyme B galactosidase which is responsible for hydrolysis of disaccharide into its monomeric units galactose and glucose. Gene Y codes for the enzyme permease which increases permeability of all to lactose and gene a codes for the enzyme transacetylase.
In switch on mechanism the regulator gene synthesizes a repressor protein and forms inducer repressor complex to transcribe the 3 structural genes and by translation all 3 enzymes are synthesized which are needed for lactose metabolism.
In the switch off mechanism in the absence of inducer the repressor binds to operator gene and RNA polymerase is not allowed to transcribe the operon and thus no translation and no synthesis of enzymes takes place.
![]()
Question 31.
What is DNA finger printing? Mention steps involved in DNA finger printing technique.
Answer:
DNA finger printing:
It is a comparison of DNA from different sources to establish the identity.
This technique involves VNTR ie variable number of tandem repeats which are different between two individuals and are used in identifying different human beings.
Steps involved in DNA finger printing:
(a) Collection of samples: A sample of blood, semen (sperm) saliva, hair follicle, skin, bone or any other tissue even us dried condition is collected.
(b) Extraction of DNA: Sample is centrifuged at 200 P PM to spin off and separate the cells. Sample of DNA is treated with REN (restriction endonuclease enzyme) to obtain fragments of DNA of different length.
(c) Separation of DNA fragments: The DNA fragments are allowed to separate on agarose gel Electro phoresis. DNA fragments are separated on the gel plate according to their density gradient where heavier DNA stands are settling at the bottom of the gel plate and lighter fragments towards the top. These DNA fragments are treated with alkaline solution to dissolve H2 bonds to produce single stranded DNA.
(d) Extraction of DNA fragments: Single stranded DNA fragments on the gel plate are extracted on to the nylon mesh or nitro cellulose membrane by the process called southern blotting technique.
(e) Tagging Radio active probes: The nylon mesh containing single stranded DNA fragments are incubated with radio active probe. Probe is a radio active synthetic DNA of known nucleotide sequence.
(f) Auto radiography: An x-ray film is placed on the nylon mesh to obtain an autoradiography print. X-ray film is then processed to obtain visible patterns of bonds of DNA which is DNA fingerprint. A comparison of two different DNA finger prints can provide information about degree of similarities and differences.
Question 32.
Mention any five goals and salient features of HGP.
Answer:
Human Genome Project (HGP) is a mega project. It has some goals and salient feature.
Goals of HGP: Some of the important goals of HGP were as follows:
- Identify all the approximately 20000 – 25000 genes in human.
- Determine the sequences of the 3 million chemical base pains which make up in databases.
- Store the information in databases.
- Improve tools for data analysis
- Transfer related technologies to other sectors such as industries.
- Address the ethical, legal, social issues that may arise from the project.
Nucleic acids are macromolecules found in the nucleus as well as cytoplasm. Nucleic acids are broadly of two types Deoxyribonucleic acid (DNA) and Ribonucleic acid (RNA). DNA acts as the genetic material in most of the organisms. RNA acts as the genetic material in some viruses. RNA functions as adapter, structured and in some cases as a catalytic molecule.
DNA: It is a long polymer of deoxyribonucleotides. It is characteristic of an organism.
Structure of Polynucleotide chain.: A nucleotide has three components a nitrogenous base, pentose sugar (ribose and case of RNA And deoxyribose for DNA) and a phosphate group.
There are two types of nitrogenous bases – purine (Adenine and Guanine) and pyrimidine’s (Cytosine, Uracil and Thymine). Cytosine is common for both DNA and RNA and thymine is present in DNA. Uracil is present in RNA at the place of thymine.
Nucleoside consists of nitrogenous base and pentose sugar through glycosidic linkage. Nucleosides of DNA are deoxyadenosine, deoxyguanosine, deoxycytidine, deoxythymidine.
When a phosphate group is linked to a 5 OH group of nucleoside through phosphodester linkage a nucleotide is formed. More nucleotides form a polynucleotide chain.
In RNA pyrimidine base found is uracil instead of thymine.
Freidrich Meischer in 1969 noticed an acidic substance present in nucleus termed as nuclein. Watson and Crick in 1953 gave the double helical structure of DNA. . .
One of the hall marks of this proposition was base pairing between the two strands of polynucleotide chains.
Chargaff found out that for a double stranded DNA the ratios between adenine and Thymine and guanine and cytosine are constant and equals one.
The salient features of double helical structure of DNA are as follows.
- It is made of two polynucleotide chains, where the backbone is constituted by sugar-phosphate.
- The two chains have anti parallel polarity. It means if one chain has the polarity 5’-3’ other has 3’-5’.
- The bases in two strands are paired through hydrogen bonds forming base pairs. Adenine forms two bonds with thymine and guanine forms three H bonds with cytosine. As a result always purine comes opposite to pyrimidine.
- The two chains are coiled in a right handed fashion. Thtch of helix is 3.4mm and there are roughly 10BP in each turn. Diameter of the helix is 0.34nm

→ Search For Genetic Material:
Transforming principle: In 1928, Frederick Griffith in a series of experiments with Streptococcus pneumonia witnessed a transformation in bacteria.
When streptococcus pneumonia bacteria are grown on a culture plate, some produce smooth shiring colonies (S) while others produce rough colonies (R). This is because the S strain bacteria have a mucous polysaccharide coat, while (R) strain does not. Mice infected with S strain (Virulent) die from pneumonia infection but mice infected with R strain do not develop pneumonia.
- S Strain – Inject into Mice – Mice die
- R Strain – Inject into Mice – Mice lives
- S Strain (heat-killed) – Inject into mice – mice live.
- S Strain (heat-killed) + R strain (live) – Inject into Mice – Mice die.
He observed that when heat-killed S strain bacteria was injected with a mixture of heat killed S bacteria and live R bacteria the mice died.
He concluded that the R strain bacteria had some how been transformed by the heat killed S strain bacteria. This transforming principle is DNA.
![]()
→ Biochemical Characterization of Transforming Principle:
Avery, Maccleod and Mccavty’s work stated the genetic material was thought to be a protein. They purified biochemical (Proteins, DNA, RNA etc) from the heat killed S cells to see which ones could transform R cells into S cells. They discovered that DNA from S bacteria caused R bacteria to become transformed.
They also discovered that protein digesting enzymes (proteases) and RNA digesting enzymes (RNases) did not affect transformation so the transforming substance was not protein or RNA. Digestion with DNase inhibited transformation suggesting that DNA caused the transformation.
Thus DNA is the hereditary material in all organisms.
→ Genetic Material is DNA:
Alfred Hershey and Martha Chase also gave experiments to prove DNA is the genetic material. They worked on viruses infecting bacteria called bacteriophages.
Bacteriophage attaches to bacteria and its genetic material enters bacterial cell. The bacterial cell treats the viral genetic material as if it was its own and manufactures more virus particles.
Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria.
They grow some viruses on medium that contained radioactive phosphorus and some others on medium that contained radioactive sulfur.
Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not.
Similarly viruses growing on radioactive sulfur contained radioactive protein but not radioactive DNA because DNA does not contain sulfur.
Radioactive phage were allowed to attach to E. coli bacteria. As the infection proceeded viral coats were removed from the bacteria agitating them in ablender. Virus particles were separated from bacteria by spinning them in a centrifuge.
Bacteria infected with viruses that had radioactive DNA were radioactive indicating that DNA was the material that passed from virus to bacteria.
Bacteria that were infected with viruses that had radioactive proteins were not radioactive. This indicates that proteins did not enter the bacteria from the viruses DNA is the enetic material that is. passed from virus to bacteria.

![]()
→ Properties of Genetic Material (DNA Vs RNA)
- It should be able to generate its replica.
- It should be chemically and structurally stable.
- It should provide for slow changes, (mutation) required for evolution.
- Protein does not fulfill the criteria hence it is not the genetic material.
- RNA and DNA fulfill the criteria.
→ RNA is Unstable:
- 2 – 0 H group present at every nucleotide (ribose sugar) in RNA is a reactive group and makes RNA liable and
easily degradable. - RNA is known as catalyst hence reactive RNA is Unstable and mutates faster.
ONA is More Stable:
- Stability is one of the properties of genetic material.
- DNA is chemically less reactive and structurally more stable when compared to RNA.
- Presence of thymine in place of uracil confers additional stability to DNA.
- Two strands being complimentary if separated by heating come together when appropriate conditions are provided,
→ Better Genetic Material (DNA OR RNA):
- Presence of thymine in place of uracil confers more stability to DNA.
- Both DNA, RNA are able to mutate.
- RNA can directly code for synthesis of proteins.
- For transmission of genetic information RNA is better.
Hence DNA is better genetic material than RNA.
![]()
→ DNA replication: The parental DNA molecule produces two identical copies of daughter DNA molecules and it is called DNA replication.
- The process of replication begins at a specific point on DNA strand called initiation site or site of origin (ORI).
- The hydrogen bonds present between two strands are dissolved by DNA polymerase I.
- The double stranded DNA is now getting separated to form single strands. This mechanism is called unzipping of DNA.
- As the double helix is separated it gets into Y shaped appearance called replication fork. Consequently on one strand (the template with polarity (3’ – 5’) the replication is continuous while on the other (polarity 5’ – 3 ’) it is discontinuous. Discontinuously synthesized fragments are joined together by DNA ligase.

→ The Experimental Proof:
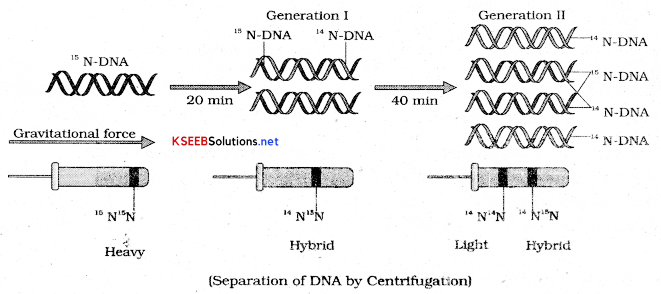
Matthew Messelson and Franklin Stahl performed the following experiment in 1958:
1. They grew E coli in a medium containing NH4 Cl is the heavy isotope of nitrogen. The result was that N was incorporated into newly synthesized DNA as well as other nitrogen containing compounds. This heavy DNA molecule could be distinguished from normal DNA by centrifugation in a calcium chloride density gradient (Cscl).
2. Then they transferred cells into a medium with normal NH4 Cl and took samples at various definite time intervals as cell multiplied.
3. Thus DNA that was extracted from culture one generation after the transfer from N to N medium had a hybrid or intermediate density. DNA extracted from the culture after another generation ie after 40 minutes was composed of equal amount of hybrid DNA and light DNA.
The experiments proved that DNA in chromosomes also replicate semi conservatively.
Transcription; Process of copying genetic information from one strand of DNA into RNA is termed as transcription. Here also the principle of complementarity governs the process of transcription except that adenosine forms base pair with uracil instead of thymine.
Transcription Unit: A transcription unit in DNA is defined primarily by three regions in DNA.
- A promoter
- Structural gene
- A terminator
Since the two strands have opposite polarity and the DNA dependent RNA polymerase also called as polymerization in only one direction that is 5’ – 3’ the strand that has polarity 3’ – 5’ – acts a template and is referred to as template strand. The strand which does not code for any thing is coding strand.
The promoter and terminator flank the structural gene in the transcription unit. The promoter is said to be located towards 5’ end of the structural gene. It is a DNA sequence that provides binding site for RNA polymerase. Terminator is located towards 3 ’ end of the coating strand and it defines the end of the process of transcription.

Transcription Unit and Gene – Cistron is a segment of DNA Coding for a Polypeptide. Structural gene in a transcription unit could be monocistronic (mostly in euparyotes) or polycistronic(bacteria or prokaryotes)
In Eukaryotes monocistronic’ structural genes are split. Coding sequences are defined as exons. Exons are interrupted by introns. Introns do not appear in mature or processed RNA.
![]()
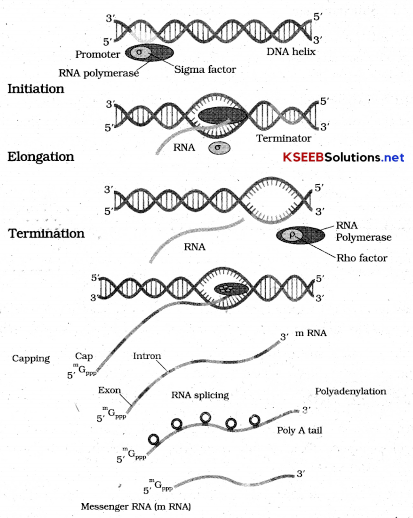
→ Types of RNA And The Process of Transcription:
In bacteria there arc major types of RNA’s mRNA tRNA and rRNA. All 3RNA’s are needed to synthesize a protein in the cell m RNA provides template, TRNA brings amino acids and reads the genetic code, or RNA’s play structural and catalytic role during translation.
There is a single DNA dependent RNA polymerase that catalyses transcription of all types of RNA in bacteria. RNA polymerase binds to promoter and initiates transcription. It facilitates opening of helix and continuous elongation. Once polymerases reaches the terminator region nascent RNA falls of and so also RNApolymerases. This results in termination of transcription.
In eukaryotes there are 3 types of RNApolymerases in the nucleus. RNA polymerase I transcribes or RNA where as RNA polymerase III is responsible for transcription of t RNA, 5 Sr RNA, SnRNA. The RNA polymerase II transcribes precursor of in RNA (the heterogeneous nuclear RNA (HnRNA).
In eukaryotes primary transcripts contains both exons and introns and is non functional. Hence it is subjected to a process called splicing where the introns are removed and exons are joined in a defined order hn RNA under¬goes additional processing called as capping and tailing. In capping an unusual nucleotide (methyl guanosine triphosphate) is added to 5’end of hn RNA.
In tailing adenylate residues (200 – 300) are added at 3’ in a template independent manner.
→ Genetic Code:
Features:
- Genetic code is universal: It is common and found in all living organisms.
- Triplet codon: Genetic code is a triplet code and is represented by three-nucleotide sequences present on m RNA strand.
- Initiator codon is AUG which codes for methionine. In case AUG is absent GUG acts as an initiator Codon which codes for amino acid valine.
- Process of translation is stopped by terminator codons UAA, UAG, UGA which do not code for any amino acid.
- Genetic code is commaless
- It is sensible
- It is non-overlapping.
→ Mutations and Genetic Code:
Mutations can be sudden and heritable change in an offspring. Mutation can be of 2 types
- Point Mutation.
- Frameshift Mutation.
| Point Mutation. | Frameshift Mutation. |
| 1. Change of singe base pair in the gene | 1. It occurs due to insertion or deletion of base |
| Ex: Sickle called anemia is due to change of pairs in the gene | Ex: Thalassemia amino acid residue glutamate to valine. |
![]()
→ TRNA the adapter molecule:
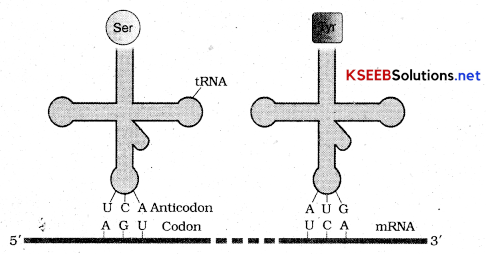
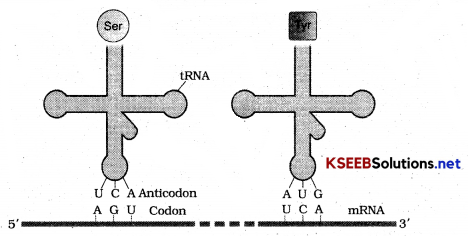
T RNA is termed as adaptor molecule as it would read the code on one hand and on the other hand would bind to specific amino acids.
Transfer RNA or T RNA has an anticodon loop that has bases complimentary to the code. It has amino acid acceptor end to which it binds to amino acids. T RNA’s are specific to each amino acid. For initiation there is initiator t RNA. It has clover leaf structure.
Translation: The process of polymerisation of amino acids to form a polypeptide is termed as translation. The process of translation takes place in the cytoplasm of the cell where ribosome is located and it takes place in 4 levels
1. Activation or charging of amino acid: Amino acids before they attach to t RNA should be activated by using ATP molecules
Amino acid + ATP- Activated amino acid.
Activated amino acid will be picked up by at RNA molecule and form amino a CY1 tRNA.
This process is called charging of amino acid.
2. Chain initiation:
- The formation of polypeptide chain is initiated by the initiator codon AUG.
- First mRNA binds to smaller subunit of ribosome ie (3os) and later on larger submit joins.
- Larger sub unit (5os) has a site (Aminoacyl site), P site peptidyl site (enzyme peptidyl transfer are and an E site or exit site.
3. Chain elongation: Ribosomal subunit shifts from one codon to another codon on the m RNA strand. T RNA carrying an amino acid shifts from A site to P site. At P site of 5os subunit peptide bond formation takes place between 2 amino acids and it is catalyzed by peptidyl transfer enzyme and elongation factos EFp EF2, EF,. chain keeps elongating.
4. Chain termination: Codons present on m RNA strand are translated by ribosomes continuously to form a long polypeptide chain until the ribosomes reach the terminator codons (UAA, UAG, UGA).
The long polyptide chain so formed is dissociated from the ribosomeand released into the cytoplasm of a cell that becomes into a type of protein.
Chain termination is characterized by release factors RF1, RF2 RF3.
Image
Lac operon concept: The lac operon concept was given by Jacob and Monod. This concept was found in bacteria.
Lac operon- consists of One regulatory gene (I gene) and three structural gene (Z, y, a) promoter gene gene Z codes for enzyme B galactosidase which is responsible for hydrolysis of disaccharide into its monomeric units galactose and glucose. Gene Y codes for the enzyme permease which increases permeability of cell to lactose and gene a codes for the enzyme transacetylasd.
In switch on mechanism the regulator gene synthesizes a repressor protein and forms inducer repressor com¬plex to transcribe the 3 structural genes and by translation all 3 enzymes are synthesized which are needed for lactose metabolism.
In the switch off mechanism in the absence of inducer the repressor binds to operator gene and RNA polymerase is not allowed to transcribe the operon and thus no translation and no synthesis of enzymes takes place.

Human Genome Project (HGP) is a mega project. It has some goals and salient feature.
→ Goals of HGP: Some of the important goals of HGP were as follows.
- Identify all the approximately 20000 – 25000 genes in humans.
- Determine the sequences of the 3 million chemical base pairs which make up in databases.
- Store the information in databases.
- Improve tools for data analyses
- Transfer related technologies to other sectors such as industries.
- Address the ethical, legal, social issues that may arise from the project.
![]()
→ Salient features of Human Genome:
Some of the salient features of HGP
- The human genome contains 3164.7 million nucleotides.
- The average gene consists of 3000 bases largest known human gene is dystrophin which has 2.4 million bases.
- Less than 2% of the genome codes for proteins.
- Repeated sequences make up a large portion of the human genome.
- Chromosome I has most genes (2968) and Y has fewest 231.
- Scientists have identified about 1.4 million locations were single base DNA differences occur in human.
→ Applications:
- Helps in diagnosing and treatment for genetic.
- DNA finger printing.
- It is a comparison of DNA from different sources to establish the identity.
- This technique involves VNTR ie variable number of tandem repeats which are different between two individuals and are used in identifying different human beings.
→ Steps Involved in DNA Finger Printing:
(a) Collection of samples: A sample of blood, semen (sperm) saliva, hair follicle, skin, bone or any other tissue even us dried condition is collected.
(b) Extraction of DNA: Sample is centrifuged at 200 or PM to spin off and separate the cells. Sample of DNA is treated with REN (restriction endonuclease enzyme) to obtain fragments of DNA of different length.
(c) Separation of DNA fragments: The DNA fragments are allowed to separate on agarose gel Electro phoresies. DNA fragments are separated on the gel plate according to their density gradient where heavier DNA stands are settling at the bottom of the gel plate and lighter fragments towards the top. These DNA fragments are treated with alkaline solution to dissolve H2 bonds to produce single-stranded DNA.
(d) Extraction of DNA fragments: Single stranded DNA fragments on the gel plate are extracted on to the nylon mesh or nitro cellulose membrane by the process called southern blotting technique.
(e) Tagging Radio active probes: The nylon mesh containing single stranded DNA fragments are incubated with radio active probe. Probe is a radio active synthetic DNA of known nucleotide sequence.
(f) Autoradiography: An X-ray film is placed on the nylon mesh to obtain an autoradiography print. X-ray film is then processed to obtain visible patterns of bonds of DNA which is DNA fingerprint. A comparison of two different DNA finger prints can provide information about degree of similarities and differences.
→ Applications of DNA Finger Printing:
- This technique is used in identification of criminals like murderers, robber rapist etc.
- It is employed in solving parental disputes.
- It is used in identifying and reunion of lost children with their parents during natural disasters like earthquake, flood etc.
- It is useful in identifying the victims of plane crash, building collapse, fire breakouts etc.
- Useful in determination of genetic diversity and evolutionary biology.
![]()
→ Salient Features of Human Genome:
Some of the salient features of HGP
- The human genome contains 3164.7 million nucleotides.
- The average gene consists of 3000 bases largest known human gene is dystrophin which has 2.4 million bases.
- Less than 2% of the genome codes for proteins.
- Repeated sequences make up a large portion of the human genome.
- Chromosome I has most genes (2968) and Y has fewest 231.
- Scientists have identified about 1.4 million locations where single-base DNA differences occur in human.