Karnataka 2nd PUC Biology Notes Chapter 6 Molecular Basis of Inheritance
Nucleic Acids:
→ Nucleic acids are large biological macromolecules formed in all living organisms. These are found in the nucleus and also in the cytoplasm of the cell. These carry genetic information from one generation to the next generation. These were first discovered by Friedrick Miescher and the term nucleic acid was coined by Altmann.
→ In eukaryotes the nucleic acids are associated with histone protein to form nucleoprotein and in prokaryotes nucleic acids are not associated with histone proteins.
Deoxyribonucleic Acid(DNA):
Occurrence: The DNA is present in all plants, animals, prokaryotes and in some viruses. In eukaryotes it is present in the nucleus, chloroplasts and mitochondria. In prokaryotes it is present in the cytoplasm.
![]()
Chemical Composition of DNA:
DNA is a macromolecuie and is made up of monomeric units [structural and functional units] called nucleotides. Chemically, each deoxy-ribonucleotide consists of 3 components namely.
- A pentose sugar
- A nitrogen base
- A phosphate group
1. Sugar: DNA consists of deoxyribose sugar which is a pentose sugar. Four carbons of a sugar namely C1, C2, C3 and C4 are joined forming a ring and the fifth carbon is present outside the ring. At the second carbon atom there is only hydrogen, no oxygen and hence it is deoxyribose sugar.
2. Nitrogen bases: There are two groups of nitrogen bases namely purines and pyrimidines.
(a) Purines: These are double ringed and heterocyclic molecules. They contain a pyrimidine ring and imidazole ring with nitrogen at 1, 3, 7 and 9 positions There are two types of purines namely adenine [A] and guanine [G].
(b) Pyrimidines: These are single ringed and heterocyclic molecules with nitrogen at 1′ and 3′ positions. There are two types pyrimidines namely cytosine [C] and thymine [T]. Thymine is present only in DNA and absent in RNA.
3. Phosphate group: It is orthophosphoric acid [H3PO4] molecule with three OH groups and one oxygen atom which is attached to the phosphorous with double bond. This phosphate group is attached to the sugar at the 5th carbon with phosphoester bond:
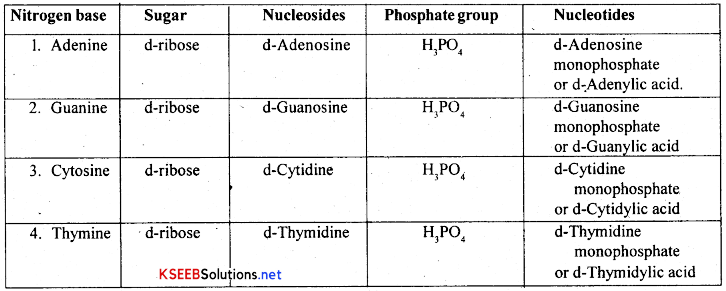
Nucleosides and Nucleotides of DNA:
The combination of a nitrogenous base and a pentose sugar is called a nucleoside A nitrogen base gets attached to a pentose sugar at carbon T with a glycoside bond. The combination of a nucleoside and a phosphate group is called a nucleotide. The phosphate is attached to the nucleoside at 5th, carbon of the sugar with Phospho-ester bond. There are four types of nucleosides and four types of nucleotides in DNA and these are as follows.
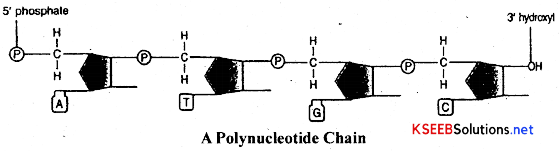
Polynucleotide strand: A long chain molecule formed by the polymerization of many nucleotides is called polynucleotide chain. The two chemical bonds formed between the phosphate group and the pentose sugars on either side are called Phospho-di-ester bonds. Two such polynucleotide strands are joined antiparallely and complementarity through hydrogen bonds forming double stand DNA molecule.
![]()
Structure of DNA-or-Watson and Crick Model of DNA:
James D. Watson and Frances H.C.Crick in 1953 proposed the model to explain the structure of DNA, based on x- ray diffraction structure by Wilkins. According to this model, DNA has the following features.
- A DNA molecule is composed of two polynucleotide strands, which are twisted right-handedly like a spiral staircase.
- Each polynucleotide strand consists of number of deoxy-ribonucleotides attached to one another by phosphodiester bonds.
- The two polynucleotide strands are joined by the specific base pairing i.e purines with pyrimidines and viceversa.
- The base pairing is specific among purines and pyrimidines. Adenine of one strand always pairs with thymine of another strand. Guanine always pairs with cytosine.
- The adenine and thymine are joined by two hydrogen bonds. The guanine and cytosine are joined by three hydrogen bonds.
- In these two strands the sugars and phosphates form the backbone and nitrogen bases that are projected towards the inner side forms axis of the molecule.
- The two strands of the DNA are antiparallel and complementary i.e One strand with 5′ to 3′ pairs with others strand, in opposite manner [3′ to 5′ manner]
- Thus in one strand the phosphate is present in the 5th carbon of sugar and at the opposite and OH is present at 3rd carbon of the sugar. In other strand the ends are reversed.
- In a DNA molecule, the amount of purines is always equal to the amount of pyrimidine ie A + G = T + CorA = T and G = C This is called Chargoff’s law or Equivalent base rule.
- Each turn in the DNA molecule consists of ten nucleotide pairs. The length of each turn is, 3.4 A°, the distance between each nucleotide in a strand is 3.4 A° the distance between two strands is 20 A°.
Packaging of DNA:
- In prokaryotes, with no well-defined nucleus, the DNA is organised in large loops held by certain positively charged proteins, in a region called nucleoid.
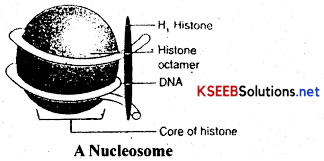
- In eukaryotes, the DNA is wrapped around a positively charged histone octamer into a structure called nucleosome.
- A typical nucleosome consists of 200 bp of DNA helix.
- The nucleosomes are the repeating units that form chromatin fibres.
- The chromatin fibres condense at metaphase stage of cell division to form chromosomes.
- The packaging of chromatin at higher level requires additional set of proteins called non-histone chromosomal (NHC) proteins.
- In a nucleus, certain regions of the chromatin are loosely packed and they stain lighter than the other regions; these are called euchromatin.
- The other regions are tightly packed and they stain darker and are called heterochromatin.
- Euchromatin is transcriptionally more active than heterochromatin.
![]()
DNA as Genetic Material:
→ The material composed of genes is called genetic material. Genes are the units of heredity. The genetic material controls the expression of ‘ characters in an organism. The genetic material has the capacity of replication and also it plays a major role in transferring characters from parental generation to the next generation.
→ In 1928, Fredrick Griffith conducted some (transformation) experiments to show that DNA is the genetic material.
→ In 1944, Oswal. T. Avery extended Griffith’s experiments and conclusively proved that DNA is the genetic material.. Avery’s experiments were purely based on the Griffith experiments.
The experiments and conclusions of Avery are as follows.
1. When a smooth strain was cultured in the medium, smooth colonies were formed.
2. When rough strain was cultured in the medium, rough colonies were formed.
3. When smooth strain bacteria are heat killed and cultured, then no colonies were observed.
4. When rough strain is mixed with heat killed smooth strain or with its DNA only and cultured, smooth colonies were observed. Thus rough strain is transformed into smooth strain.
5. When heat killed smooth type bacteria and DNAase enzyme [which digests the DNA] are mixed with rough type bacteria and then cultured, no smooth colonies were observed. This showed that DNA was responsible for trans forming rough type into smooth type and thus it shows that DNA acts as genetic material. This process of passing of hereditary molecule from the heat killed smooth type to the rough type and converting to ‘R’ type bacteria to ‘S’ type bacteria is called transformation.
Hershey-Chase Experiment:
→ The proof for DNA as the genetic material came from the experiments of Alfred Hershey and Martha Chase (1952), who worked with bacteriophages.
→The bacteriophage on infection injects only the DNA into the bacterial cell and not the ! protein coat; the bacterial cell treats the viral DNA as its own and subsequently manufactures more virus particles.
→ They made two different preparations of the phage; in one, the DNA was made radioactive with 32P and in the other, the protein coat was made radioactive with 35S.
→ These two phage preparations were allowed to infect the bacterial cells separately.
→ Soon after infection, the cultures were gently agitated in a blender to separate the adhering protein coats of the virus from the bacterial cells.
→ The culture was also centrifuged to separate the viral coat and the bacterial cells.
→ It was found that when the phage containing radioactive DNA was used to infect the bacteria, its radioactivity was found in the bacterial cells (in the sediment) indicating that the DNA has been injected into the bacterial cell.
→ So, DNA is the genetic material and not proteins.

![]()
Functions of DNA:
- DNA stores and transmits genetic information from parents to off springs.
- It controls all cellular activities of the cell.
- It has the capacity to undergo mutation leading to variations.
- It acts as a template for the synthesis of different types of RNAs.
- It directs the synthesis^of proteins and helps in the growth and development.
- It directs the synthesis of specific enzymes of RNA.
Ribonucleic Acid RNA:
Occurrence: It is a type of nucleic acid present in both the prokaryotes and eukaryotes. It is mainly found in the cytoplasm and a little in nucleus of eukaryotes. It is made up of a single polynucleotide strand.
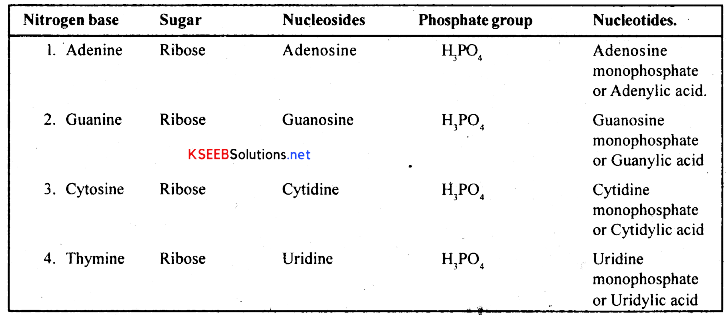
Chemical composition: RNA is a biological macromolecule made up of four types of ribonucleotides. Each ribonucleotide consists of apentose sugar, a nitrogen base and a phosphate group.
1. Sugar: RNA has ribose sugar, which is a pentose sugar. Four carbons are arranged in the ring form with oxygen bridge between carbon 1 ’ and 4’. The fifth carbon is present out side the ring. There is an (OH) at the second carbon.
2. Nitrogen bases:- These are of two groups namely purines and pyrimidines. Purines are double ringed and heterocyclic. These are of two types namely Adenine [A] and Guanine [G]. These are structurally similar to those of DNA. Pyrimidines are of two types namely Cytosine [C] and Uracil [U]. Uracil is present only in RNA and absent in DNA.
3. Phosphate group:- It is orthophosphoric acid [H3P04] molecule with phosphorus in the center and QH groups on three sides and oxygen on one side with double bonds.
Nucleosides and Nucleotides of RNA: There are four types nucleosides and four types of nucleotides in RNA. They are as follows.
![]()
Polynucleotide strand:
A long chain formed by the polymerization of many nucleotides is called Polynucleotide strand. This forms the RNA molecule.
Types of RNA: There are two categories of RNAs namely genetic RNA and non-genetic RNA.
- Genetic RNA:- It is the RNA which acts as genetic material and carries hereditary characters from one generation to another. It is found in viruses like TMV, [ssRNA], Wound tumour virus [dsRNA] HIV, Poliomyelitis, [ssRNA] Reo virus [ds RNA]
- Non-genetic RNA:- It is the RNA involved in protein synthesis. It is of three types namely RNA, m-RNA and t-RNA. All these 3 types of RNAs are synthesized by DNA
(a) Ribosomal RNA or r-RNA: It is the ribosomal RNA. It forms the structural component of ribosome and it accounts for 80% of the total RNA of the cell. It is a single polynucleotide strand with number of curves and coiling at some regions. It is rich with guanine and cytosine. This r-RNA combines with specific type of proteins forming ribosome.
Functions:
- It helps in the binding of mRNA and tRNA to the ribosome during protein synthesis.
- It is involved in the formation of ribosome.
- It acts as an enzyme, ribozyme in peptide bond formation between aminoacids.
(b) Messenger RNA or Informational RNA or mRNA:
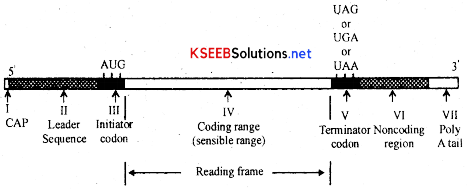
→ It is the messenger RNA synthesized by DNA on its templates. It is the largest RNA [900-1500 nucleotide] of all the types. It accounts for about 5% of the total RNA in the cell It was discovered by Volkin and the term mRNA was coined by Jacob and Monad. The life span of mRNA is 2 minutes [1 to 4 hours in Eukaryotes]
→ Structurally eukaryotic mRNA has a guanosine nucleotide cap at 5 end. This cap protects the mRNA from being digested by enzymes. It is followed by a short segment of leader sequence which do not code for aminoacids [nOn-coding region 1 ] [200 Nb]. This region helps in binding of mRNA to ribosome it is followed by Coding region which has message for the sequence of aminoacids in the proteins to be synthesised. This coding region starts with initiator codon-AUG and ends with any one of the three non-sense codons [UAA, UAQ UGA].
→ It is followed by a short non coding region II called trailer sequence. Next to the trailer sequence there is poly ‘A’ tail at 3’ end. ‘A’ tail [AAA] is absent in mRNA of prokaryotes. There are two types mRNAs namely monocistronic and polycistronic mRNA. The mRNA which is transcribed from only one cistron or gene is called monocistronic mRNA and is present in eukaryotes. The mRNA which is transcribed from many cistrons or genes is called polycistronic mRNA The polycistronic mRNA are commonly found in prokaryotes.
Functions: It carries message from DNA to cytoplasm in the form of triplet codons for the synthesis of proteins.
![]()
(c) Soluble RNA or transfer RNA [tRNA]:
It is the transfer RNA synthesized by the DNA. It is the smallest RNA with 80 nucleotides. It accounts for about 15% of the total RNA of a cell. It is primarily a single stranded molecule but the nucleotides at certain regions pairs with one another to form double stranded stems. It looks like a clover leaf and forms a secondary structure. This secondary structure was first discovered and described by Robert Holley who got Nobel prize for the same.
Secondary structure of tRNA consists of four arms namely acceptor arm, DHU arm, anticodon, arm and T Ψ C arm Each arm has a stem and a loop but the acceptor arm is loopless. The stem has paired bases and the loop has impaired bases. The four arms are as follows.
1. The acceptor arm: This is formed by 7 base pairings between nucleotides at the 5’ and 3′ ends of tRNA. Beyond the base pairings there extend three unpaired nucleotides CCA at 3’end. The aminoacid gets attached to the OH group of last adenine nucleotide. Hence this 3’ end is called aminoacid binding site.
2. DHU or- Dihydrouridine arm: It is towards 5′ end of the molecule after the acceptor arm. It has 4 base pairs at its stem and 10 unpaired nucleotides at its loop. It has art unusual pyrimidine called Dihydrouridine. This arm has a specific charging enzyme that catalyses the union of specific amino acid to tRNA molecule. Hence it is also called aminoacid activating site [enzyme site]
3. The anticodon arm: It is opposite to the acceptor arm. It has 5 base pairs in its stem and 7 unpaired base pairs. Three unpaired bases of the loop forming anticodon or NODOC which is responsible for recognizing and binding to the codon of mRNA.
4. T Ψ C or pseudo uridine arm: It is opposite to DHU arm. It has 5 Bp in stem and 7 unpaired nucleotides in the loop. It contains unusual nucleotides like pseudouridine, dimethyl guanosine and inosine. This arm is responsible for recognition of ribosomes during protein synthesis. Hence called ribosome recognition arm or site. Variable arm: It is small extra arm present in between the anticodon arm and T Ψ C arm in some tRNAs. Its function is not yet known.
Functions:
- It transfers a specific aminoacid to the site of protein synthesis.
- Protein synthesis makes use of about 20 types aminoacids. So there are 20 different tRNAs in a cell to carry 20 types of aminoacids.
- It carries information at its anticodon.
![]()
Differences between DNA and RNA:
| DNA | RNA |
| 1. It is usually genetic material. | 1. It is rarely genetic material |
| 2. It is double stranded and helical | 2. It is single stranded and non- helical |
| 3. It has a deoxv ribose sugar | 3. It has a ribose sugar |
| 4. Nitrogen bases are AGC and T | 4. Nitrogen bases are AGC and U |
| 5. It replicates by itself | 5. It is synthesized by DNA |
| 6. It occurs in chromosomes | 6. It occurs in cytoplasm and nucleolus |
| 7. Amount of purines is equal to that of pyrimidines. | 7. It is unequal |
| 8. Adenine pairs with thymine | 8. Adenine pairs with uracil |
| 9. It is of one functional type | 9. It is of four type |
Characteristics of Genetic Material:
→ A molecule that can act as genetic material must have the fol lowing properties :
- It should be able to generate its replica.
- It should be chemically and Structurally stable.
- It should provide the scope for slow changes (mutation) that are necessary for evolution.
- It should be able to express itself in the form of Mendelian characters.
→ Nucleic acids i.e., DNA and RNA can replicate, but not protein.
→ The predominant genetic material is DNA, while few viruses like tobacco mosaic virus have RNA as the genetic material.
→ The 2′- OH group in the nucleotides of RNA is a reactive group and makes RNA easily degradable; RNA as a catalyst is also more reactive and hence DNA has the property to be the genetic material.
![]()
Replication Of DNA:
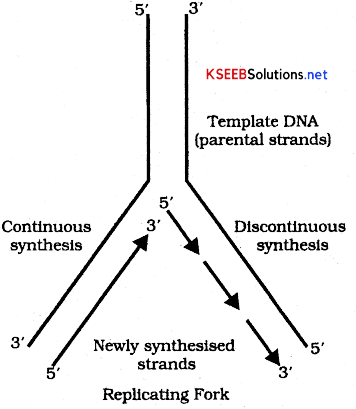
Semi-conservative mode of DNA replication: The process of formation of exact copy of DNA is called replication of DNA. It takes place by semi conservative method. It means one strand of parent molecule is conserved in the daughter molecule of DNA. It is experimentally proved by Meselson and Stahl. According to Watson and Crick DNA replication is as follows.
1. Requirements: DNA replication needs DNA templates, DNA unwindase RNA primer, RNA polymerase, DNA polymerase I and III, DNA lygase and Deoxy ribonucleotides [d-ATP, d- CTP, d-GTP. d-TTP].
2. Unwinding of DNA: Unwinding of DNA takes place from a specific site called Ori site. Here the two strands of DNA start unwinding and get separated with the help of Unwindase enzyme or Helicase. The two separated strands of DNA look like ‘Y’ shape and this is called replication fork. The separated DNA strands act as templates for the synthesis of new strand.
3. Synthesis of RNA primer: RNA primer is a short segment of RNA [10 nucleotides], synthesized on DNA template with the enzyme DNA polymerase III cannot directly initiate the synthesis the polymerization of DNA nucleotides on the template strand.
4. Formation of leading and lagging strands: Formation of new DNA strands by DNA polymerase enzyme can occur only in 3′ to 5’ direction. Synthesis of new complementary strands takes place on both the templates in opposite directions. New complementary strand that is formed towards the replication fork on parental strand is continuous and is called Leading strand or continuous strand. The synthesis of other new complementary strands from the replication fork occuring discontinuously on parental strand is called lagging strand or discontinuous strand. This lagging strand will have small segments of RNA primer to which DNA nucleotides are joined. These are called Okazaki fragments.
5. Formation of separation of daughter DNA molecule: In this step RNA primers are removed by the DNA polymerase 1 [Korenberg enzyme] and the gaps are filled up by the DNA nucleotides -by the same enzyme. Then short segments of DNA get joined by DNA lygase forming a continuous strand of DNA. Thus each daughter DNA molecule will have one old parental strand and one new stand and hence it follows semi-conservative method of DNA replication.
![]()
Transcription:
Transcription Unit
A transcription unit in DNA is defined primarily by the three regions in the DNA:
- A Promoter
- The Structural gene
- A Terminator
→ There is a convention in defining the two strands of the DNA in the structural gene of a transcription, unit. Since the two strands have opposite polarity and the DNA-dependent RNA polymerase also catalyse the polymerisation in only one direction, that is 5′ → 3′, the strand that has the polarity 3’→ 5′ acts as a template, and is also referred to as template strand. The other strand which has the polarity (5’ → 3′) and the sequence same as RNA (except thymine at the place of uraciil), is displaced through transcription. Strangely, this strand (which does not code for anything) is referred to as coding strand. All the reference point while defining a transcription unit is made with coding strand. To explain the point, a hypothetical sequence from a transcription unit is represented below:
3’ – ATGCATGCATGCATGCATGCATGC-5′ Template Strand
5′ – TACGTACGTACGTACGTACGTACG-3′ Coding Strand
Can you now write the sequence of RNA transribed from the above DNA?
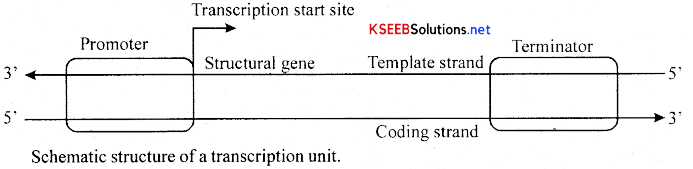
→ The promoter and terminator flank the structural gene in a transcription unit. The promoter is said to be located towards 5′ – end (upstream) of the structural gene (the reference is made with respect to the polarity of coding strand). It is a DNA sequence that provides binding site for RNA polymerase, and it is the presence of a promoter in a transcription unit that also defines the template and coding strands. By switching its position with terminator, the definition of coding and template strands could be reversed. The terminator is .located towards 3′ – end (downstream) of the coding strand and it usually defines the end of the process of transcription. (Fig). There are additional regulatory sequences that may be present further upstream or downstream to the promoter.
→ Transcription Unit and the Gene:
A gene is defined as the functional unit of inheritance. Though there is no ambiguity that the genes are located on the DNA, it is difficult to literally define a gene in terms of DNA sequence. The DNA sequence coding for tRNA or rRNA molecule also define a gene. However by defining a cistron as a segment of DNA coding for a polypeptide, the structural gene in a transcription unit could be said as monocistronic (mostly in eukaryotes) or polycistronic (mostly in bacteria or prokaryotes). In eukaryotes, the monocistronic structural genes have interrupted coding sequences – the genes in eukaryotes are split. The coding sequences or expressed sequences are defined as exons. Exons are said to be those sequence that appear in mature or processed RNA. The exons are interrupted by introns. Introns or intervening sequences do not appear in mature or processed RNA. The split-gene arrangement further complicates the definition of a gene in terms of a DNA segment.
→ Inheritance of a character is also affected by promoter and regulatory sequences of a structural gene. Hence, sometime the regulatory sequences are loosely defined as regulatory genes, even though these sequences do not code for any RNA or protein.
→ Synthesis of messenger RNA on DNA template in the presence of RNA polymerase enzyme is called transcription. Following events occur during this process.
![]()
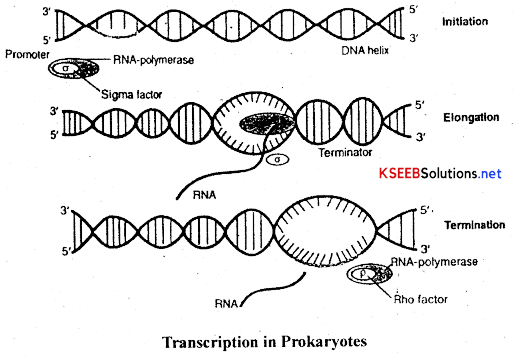
(a) Transcription in prokaryotes:
- In prokaryotes, the structural genes are polycistronic and continuous.
- In prokaryotes, there is a single DNA – dependent RNA polymerase, that catalyses the transcription of all the three types of RNA (mRNA, tRNA, rRNA).
- RNA polymerase binds to the promoter and initiates the process along with certain initiation factors(α).
- It uses ribonucleoside triphosphates (also called ribonucleotides) for polymerisation on a DNA template following complementarity of bases.
- The enzyme facilitates the opening of the DNA-helix and elongation continues.
- Once the RNA polymerase reaches the terminator, the nascent RNA falls off and the RNA polymerase also separate. It is called termination of transcription and is facilitated by certain termination factors (ρ).
- In prokaryotes, the mRNA synthesised does not require any processing to become active and both transcription and translation occur in the same cytosol. Translation can start much before the mRNA is fully transcribed, i.e., transcription and translation can be coupled.
(b) Transcription in Eukaryotes
- In eukaryotes, the structural genes are monocistronic and ‘split’.
- They have coding sequences called exons that form part of mRNA and non-coding sequences, called introns, that do not form part of the mRNA and are removed during splicing.
- In eukaiyotes, there are atleast three different RNA- polymerases in the nucleus, apart from the RNA polymerase in the organelles, which function as follows:
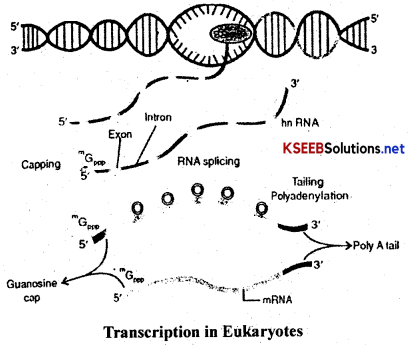
RNA polymerase I transcribes rRNAs (26S, 18S and 5.8S)
RNA poymerase II transcribes the precursor of mRNA (called as heterogenouus nuclear RNA (hnRNA)) and
RNA-polymerase III catalyses transcription of tRNA.
- The primary transcript contains both exons and introns and it is subjected to a process, called splicing, where the introns are removed and the exons are joined in a definite order to form mRNA.
- The hnRNA undergoes two additional processes called ‘capping’ and ‘tailing’.
- In capping, methylguanosine triphosphate is added to the 5’end of hnRNA.
- In tailing, adenylate residues (about 200-300) are added at the 3’end of hnRNA.
- The fully processed hnRNA is called mRNA and is released from the nucleus into the cytoplasm.
![]()
Genetic Code:
Genetic code can be defined in the following ways.
(A) It can be defined as the code language of the gene.
(B) It can be defined as biological dictionary employed by all living organisms to convert the language of genes into the language of proteins.
(C) The relation between the sequence of nucleotides in the DNA molecule and the sequence of amino acids in the protein produced by it.
(D) Message present in mRNA molecule in the form of a specific sequence of nucleotides. This was discovered by Marshal Nirenberg, Holley and Har Govind* Khorana.
The information for all proteins or enzymes is present in DNA in the form of ATGC nucleotides. This information is transcribed in to the mRNA in the form of AUGC bases. The sequence of these nucleotides decides the sequence of aminoacids in a protein. There are 20 different aminoacids in a cell. All these are necessary for the production of proteins. But DNA or RNA has only four types of nucleotides to select 20 types of aminoacids. The concept regarding possible chances of selecting the amino acids by the nucleotides in the form of codons are as follows.
1. Singlet codon system or monoplet system: According to this, a single nucleotide of mRNA forms a codon for one aminoacid. Thus it could code for four kinds of aminoacids only. But it is not possible to code for the other 16 types of aminoacids. So it is rejected.
2. Doublet codon system or duplet system: According to this, two nucleotides of mRNA form . a codon for one aminoacid. Thus 16 codons [4*4] are possible and they could code for 16 types of amino acids only and it could not answer for the remaining four kinds of aminoacids. So it is also rejected.
3. Triplet codon system: According to this three nucleotides of mRNA form a codon for one aminoacid. Thus, 64 codons are possible [4 × 4 × 4] to code for 20 types of amino acids. Among these, 61 codons code for aminoacids and are called functional or sensible codons. The other three codons, do not code for aminoacids and hence called non-sense codons. However they are required for the termination of the synthesis of polypeptide chain and hence they are called terminator codons.
According to H.GKhorana, the sequence of 3 nucleotides of DNA which codes for one aminoacid is called triplet code The sequence of 3 nucleotides of mRNA which codes for one aminoacid is called triplet codon. The code and codon of the same amino acid are complementary to each other.
DNA ——— ATG GAT CAT ACC ——— code
mRNA ——— UAC CUA GUA UGG ——— COdon
RNA ——— AUG GAU CAU ACC ——— Anticodon.
The various codons of mRNA and aminoacids selected by them are as follows.
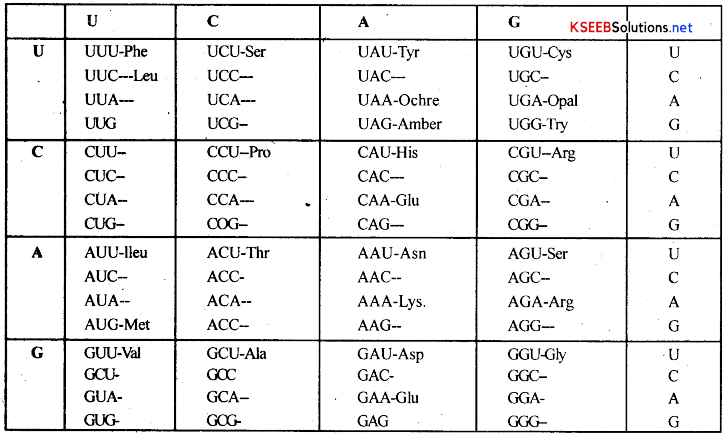
AUG = Initiator codon
AAA = Lysine
UUU = Phenyl alanine
GGG = Glycine
CCC = Proline
UAA, UAG and UGA = Terminator codons
![]()
Characters of Genetic code:
1. Genetic code is universal: It means a particular triplet codon codes for same aminoacid in all living organisms, e.g; AUG codes for methionine in all living organisms, plants and animals.
2. Genetic code is triplet in nature:- It means set of 3 nucleotides of mRNA will code for one aminoacid.
3. Genetic code is degenerative: It means a single aminoacid may be coded by more than one codon, e.g; Leucine is coded by 6 codons such as UUA, UUG CUU, CUC, CUA and CUG [In these codons the first two letters of codon and anticodon codes for the same amino acid irrespective, of the 3rd base. This is wobble hypothesis and this was proposed by Crick. The 3rd base is wobble base]
4. Genetic code is non-overlapping: When the triplet codons are read from one end of mRNA, they should be read as units of 3 nucleotides. A nucleotide of one codon does not become part of the next codon.
5. Genetic codon has initiator codon: Initiator codon always initiates the protein synthesis and is always AUG. In the absence of AUG, GUG initiates the protein synthesis.
6. Genetic code has terminator codons:- These codons do not code for any aminoacids but they terminate or stop the synthesis of polypeptide chain during protein synthesis. They include UAA, UAQ UGA.
7. Genetic code is comma, less: It means coding of aminoacid is continuous one after the other during the synthesis of polypeptide chain.
8. Genetic code has co-linearity: The sequence of nucleotides on DNA or mRNA has a relationship with the sequence of aminoacids in a polypeptide chain.
tRNA – The Adaptor Molecule
From the very beginning of the proposition of code, it was clear to Francis Crick that there has to be a mechanism to read the code and also to link it to the amino acids, because amino acids have no structural specialities to read the code uniquely. He postulated the presence of an adaptor molecule that would on one hand read the code and on other hand would bind to specific amino acids. The tRNA, then called sRNA (soluble RNA), was known before the genetic code was postulated. However, its role as an adaptor molecule was assigned much later.
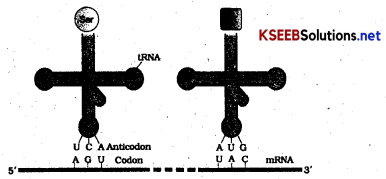
tRNA has an anticodon loop that has bases complementary to the code and it also has an amino acid acceptor end to which it binds to amino acids tRNAs are. specific for each amino acid (Fig.) For initiation, there is another specific tRNA that is referred to as initiator tRNA. There are no tRNAs for stop codons. The secondary structure of tRNA has been depicted that looks like a clover-leaf. In actual structure, the tRNA is a compact molecule which looks like inverted L.
![]()
Translation:
The process of translation of nucleotide language of mRNA into aminoacid language by tRNA is called translation. This process occurs in the cytoplasm of the cell both in prokaryotes and eukaryotes. The process of translation involves the following steps.
1. Activation of aminoacids
2. Initiation of polypeptide chain.
3. Elongation of polypeptide chain.
4. Termination of polypeptide chain.
1. Activation of aminoacids: Amino acids are present in the cytoplasm and are in inactive form These are combined with ATP in presence of an enzyme aminoacyl synthetase and Mg++ ions forming activated aminoacid.
![]()
This activated amino acid then combines with 3 end of tRNA forming aminoacyl-tRNA complex , or charged RNA and AMP is released.
2. Initiation of polypeptide chain: In this process, mRNA gets attached to 30s ribosomal subunit. Now the tRNA carrying activated aminoacid, methioninp gets attached to the first codon of mRNA. It takes place in presence of GTP and initiation factors [IF1 IF2 IF3]. This is the initiation complex. Now 50s ribosomal sub-unit combines with 30s ribosomal unit forming 70s ribosome. The first tRNA-aminoacid complex is now at ‘P’ site of ribosome.
3. Elongation of polypeptide chain: The ‘A’ site of 50s ribosome is now free and so the second tRNA with second activated aminoacid is get attached at ‘A’ site. A peptide bond is formed between 1st and 2nd aminoacid in presence of peptidyl transferase at ‘P’ site. Now the ribosome moves a distance of one codon so that the ‘A’site become vacant. Meanwhile the 3rd tRNA with 3rd aminoacid gets attached to ‘A’ site. Again the ribosome moves on mRNA by one codon and bonding takes place between 2nd and 3rd aminoacid at ‘P’ site. 1st tRNA is released through ‘E’ site. This process continues till all the codons on mRNA are completely read. Thus polypeptide chain elongates along the mRNA.
4. Termination and release of polypeptide chain: The process of protein synthesis continues till the arrival of terminator codon on mRNA at ‘A’ site. The terminator codon may be UAA, UAG and UGA. These codons do not code for any aminoacids. During this process the | enzyme peptidyl synthetase catalyses the cleavage of polypeptide chain from the last tRNA. Here releasing factors RF1 RF2 and RF3 are involved. Thus the polypeptide chain is finally released.
- The speed of transcription is 30 nucleotides per second in bacteria.
- The speed of translation 20 aminoacids per second in bacteria.
![]()
Genetic Control of Protein Synthesis or Gene Regulation of Gene Action:
DNA consists of number of genes so that they could produce all types of proteins at all the times. l But all proteins are not produced at a time in the cells of the body, as they are not needed at the same time of growth. Thus the production of proteins and action of protein producing genes are regulated. There are two groups of genes namely structural genes and control genes. Structural genes are involved in the production of mRNA for proteins. Control genes control the production mRNA from the structural genes. This composite unit of structural genes and control genes is called Operon. This concept was first proposed by Jacob and Monad with regard to lactose catabolism in. E.coli and it is called lac operon.
Elements or components of the lac operon:
Lac operon complex has the following elements.
1. Structural genes: These are the genes involved in the synthesis of mRNAto produce a protein. These genes occur close to one another, hence called clustered genes and are designated as z, y, a.
2. Control genes: These include operator, promoter and regulator genes.
(a) Operator gene: It is present in between the promoter and structural genes. This gene regulates the action of structural genes by controlling the activity promoter gene. It is the site of attachment of repressor protein. It is designated as ‘O’
(b) Promoter gene: It is present just beside the operator gene and is designated as ’P’. It is that place of gene where RNA polymerase enzyme binds. This enzyme is required to initiate the synthesis of mRNA. At this place DNA has TATAAT nucleotides in prokaryotes and this forms Pribnow box. In eukaryotes TATA nucleotides are present and this forms Hognese box.
(c) Regulator gene: It is present beside but little bit away from the promoter gene and is designated as ‘R’ It codes for regressor protein which could bind at operator and blocks it. So it regulates the movement of RNA polymerase enzyme and thus takes control over the action of operator gene.
Lac-Operon in Escherichia Coli [E.COLI]:
Regulation of gene action is well studied in E.coli bacteria with regard to utilization of lactose by the bacteria. This is an example for inductive operon because, the gene expression is turned on by the addition of lactose [Inducer] into the medium.
Utilization of lactose in E.coli needs three enzymes namely β-galactosidase, β-galactoside permease and β-galactoside transacetylase. These are produced by z, y and a structural genes respectively in presence of RNA polymerase enzyme. This enzyme is bound to the promoter region of DNA. This enzyme has to move along the structural genes to initiate the synthesis of mRNA for these 3 enzymes. ‘
The mechanism of Lac Operon can be studied under two steps namely.
- Switched OFF mechanism and
- Switched ON mechanism.
1. Switched OFF mechanism: When lactose is absent in the medium, the regulator gene produces repressor protein which binds with the operator gene. The repressor protein is an allosteric protein with 2 specific sites. One is active site with which it binds to operator gene and another is effecter site at which the lactose molecule can bind. Thus movement of RNA polymerase on the structural genes is blocked. So there is no synthesis of mRNA from the structural genes z, y and a. So there is no synthesis of enzymes. This is called Switched OFF mechanism.
2. Switched ON mechanism: When lactose is added to the culture medium some of its molecules enter into the bacterial cell and one of them binds itself with repressor. It induces the repressor protein to undergo structural change and makes it inactive. This inactive repressor becomes detached from the operator region. Now the RNA polymerase moves along the DNA and as a result the structural genes z, y and a, produce mRNA. This mRNA synthesis 3 enzymes, which are necessary for lactose metabolism. This is called switched ON- mechanism.
![]()
Human Genome Project [HGP]:
It is an international project started in 1990 and proposed to be completed by 2005,. It was meant for acquiring complete knowledge of organization, structure of function of human genome.
Goals of HGP:
- To determine complete nucleotide sequence of DNA of each chromosome.
- To study the structure, organization and function of human DNA which consists 3 billion nucleotide base pairs.
- To identify the location and structure of defective genes.
- To identify and cure genetic diseases.
- To get genetic and physical maps of human DNA.
- To determinate the functions of different genes.
Methodologies:
→ The methods involved two major approaches. One approach focused on identifying all the genes that are expressed as RNA (referred to as Expressed Sequence Tags (ESTs)). The other is simplyequencing the whole set of genome that contained all the coding and non-coding sequence, and later assigning different regions in the sequence with functions ( a term referred to as Sequence Annotation). For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes and cloned in suitable host using specialised vectors. The cloning resulted into amplification of each piece of DNA fragment so that it subsequently could be sequenced with ease. The commonly used hosts were bacteria and yeast, and the vectors were called as BAC (bacterial artificial chromosomes), and YAC (yeast artificial chromosomes).
→ The fragments were sequenced using automated DNA sequencers that worked on the principle of a method developed by Frederick Sanger. These sequences were then arranged based on some overlapping regions present in them. This required generation of overlapping fragments for sequencing. Alignment of these sequences was humanly not possible. Therefore, specialised computer based programs were developed (Fig). These sequences were subsequently annotated and were assigned to each chromosome. The 24 human chromosomes – 22 automsomes and X and Y – were sequenced. Another challenging task was assigning the genetic and physical maps on the genome. This was generated using information on polymorphism of restriction endonuclease recognition sites, and some repetitive DNA sequences known as microsatellites.
Achievements of HGP:
HGP got success in the following aspects.
- This project identified nearly 30,000 to 35,000 genes of human DNA.
- They identified 3164.7 million nitrogen bases in the human genome
- They identified the size and number of bases in genes. The average gene has 3000 bases and the largest human gene, dystrophin has 2.4 million bases.
- They identified the fact that all human beings are 99.9% identical with each other and only 0.1 % are different.
- They identified that the chromosome-1 has maximum number of genes ie 2968 genes and y chromosome has the lowest number of genes i.e. 231.
- They identified that only 2% of the genome codes for protein and remaining 98% DNA remains functionless. This non-functional DNA is called Junk DNA.
![]()
Application of HGP:
- It helps in the identification of defective genes that cause genetic diseases.
- It helps to prepare modern drugs and to understand their action.
- It helps to invent new methods of medical treatment.
DNA Finger Printing or Genetic Finger Printing or DNA Profiling:
A technique to identify a person on the basis of his or her DNA specificity is called DNA finger printing. Or the regions on DNA detected by DNA probes in the form of autoradiographic bands are called DNA finger prints. Or The process of matching a short piece of DNA-bf victim with the DNA of suspect to identify the involvement of the suspect in criminal cases is called DNA finger printing.
This was first developed by Alec Jeffry. In India it was developed by Dr.Lalji Singh and Dr.V.K.Kashyap at center for Cellular and Molecular biology, Hyderabad.
Steps of DNA finger printing:
It involves the following steps.
1. Collection of sample: Samples of blood, semen, vaginal debris, hair root, bone marrow, Skin or any other tissues are collected from the place where a crime has taken place. [If the DNA of the sample is too small then many copies of this DNA can be obtained by the process called polymerase chain reaction (PCR)].
2. Extraction and fragmentation of DNA: The sample material is treated with chemicals to obtain pure DNA. The DNA of the sample is then treated with restriction endonucleases to obtain DNA fragments which are of different lengths. Some of the fragments of DNA have repeated sequences of nucleotides and are called variable number of tandem repeats [VNTR], These VNTR’s specific to a person.
3. Separation of DNA fragments: DNA fragments are separated according to their length and are arranged on Electrophoretic gel slab by a technique called gel electrophoresis. After this, DNA fragments are arranged in a band pattern.
4. Splitting and transferring DNA by Blotting: DNA fragments on the gel slab are treated with an alkali solution to make them into a single stranded DNA. Then, these are transferred to nylon or nitrocellulose sheet so that the DNA strands become bound to the membrane exactly in the same pattern as they were in the gel. This type of transfer is called southern blotting technique, in honour of the scientist who developed the technique.
5. Attachment of radioactive probes: Nylon sheets having single stranded DNA fragments is immersed in a solution containing radioactive DNA probes. [DNA probes are short, synthetic segments of single stranded DNA which are labeled with radioactive isotopes] DNA probes bind or hybridize with specific sequences ofDNA strands present on the nylon membrane. Unhybridized DNA probes are washed off.
6. Autoradiograph: This nylon membrane is exposed to X-ray film, is then developed to make the bands visible and to detect radioactive pattern. Only hybridized DNA probes appear dark. This photograph is called autoradiograph. The position and number of bands
in autoradiograph is specific to person and vary from person to person like finger prints. This forms the DNA finger print.
7. Comparison: This DNA finger print is then compared with such DNA finger print of suspects. If the DNA finger prints of the sample and suspect is found similar, then it could be concluded that the suspect is involved in the crime.
![]()
Applications of DNA finger printing:
- It helps in solving the disputed parentage by comparing the DNA finger prints of child, mother and father.
- It helps to reunite the lost children to their respective parents.
- It helps to identify criminals.
- It helps to identify the immigrants who have crossed the borders of the country.
- It helps to show the connecting links between the different groups of animals, e.g. Human and apes.